Great job!
Archivo de la categoría: Sin categoría
A new framework to estimate evolutionary parameters from protein sequences: ProteinEvolverABC
There is a need for evolutionary frameworks to study protein evolution. ProteinEvolverABC aims to help with this by estimating substitution and recombination rates from protein multiple sequence alignments. https://academic.oup.com/bioinformatics/article/38/1/58/6358715

Congrats to Catarina Branco for the «Graduate Student Excellence Award» by the SMBE 2021!!
Catarina Branco was awarded with “Graduate Student Excellence Award” by SMBE 2021. To appear in https://www.smbe.org/smbe/AWARDS/SMBEGraduateStudentExcellenceAwards.aspx Congrats Catarina!!
https://www.farodevigo.es/gran-vigo/2021/08/21/universidad-vigo-mejores-promesas-mundo-56401055.html
Congrats Roberto del Amparo for the PhD Thesis with the highest score!! Well done!!
Roberto obtained his PhD last 22 July (2021) with the highest score. The thesis was about modeling protein evolution. Good job Roberto!!
Congrats to Roberto del Amparo for the Prize to the best Poster of the 4th Annual Meeting of the CINBIO!!
Roberto won the Prize to the most outstanding Poster of the 4th Annual Meeting of the CINBIO. The poster presents our work on new substitution models of HIV-1 protein evolution. Congrats Roberto!!

Open! job: Bioinformatician in protein modeling
See section Vacancies https://cme.webs.uvigo.es/vacancies/
Influence of Paleolithic range contraction, admixture and long‐distance dispersal on genetic gradients of modern humans in Asia
We found that several evolutionary and environmental processes affected the current (east-west) Asian genetic gradient of modern humans. Published in Molecular Ecology, https://onlinelibrary.wiley.com/doi/abs/10.1111/mec.15479

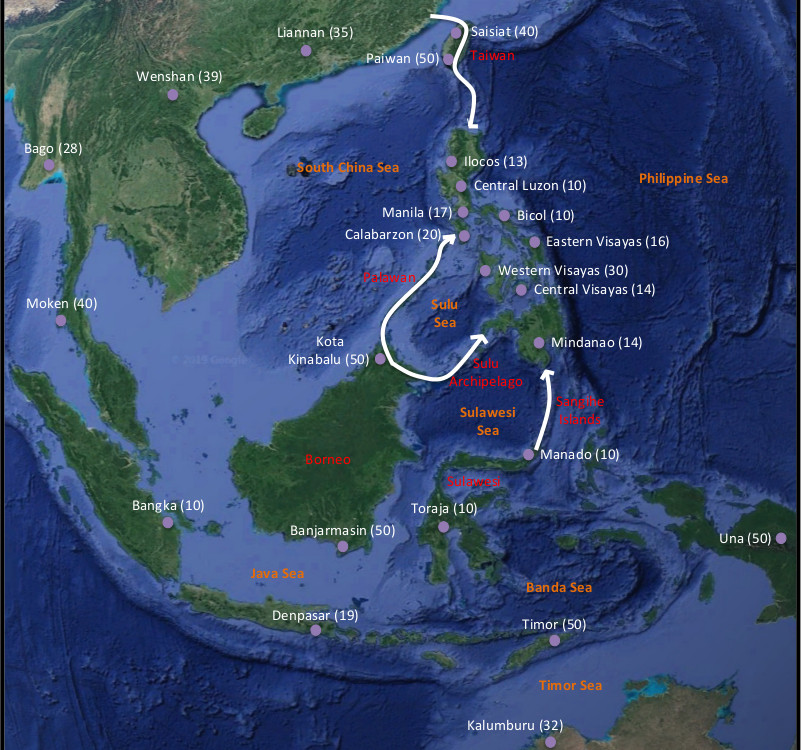
The Influence of Heterogeneous Codon Frequencies along Sequences on the Estimation of Molecular Adaptation
We found that ignoring the variation of codon frequencies among sites biases the estimation of molecular adaptation (dN/dS). Published in Bioinformatics, https://academic.oup.com/bioinformatics/article-abstract/36/2/430/5532222?redirectedFrom=fulltext
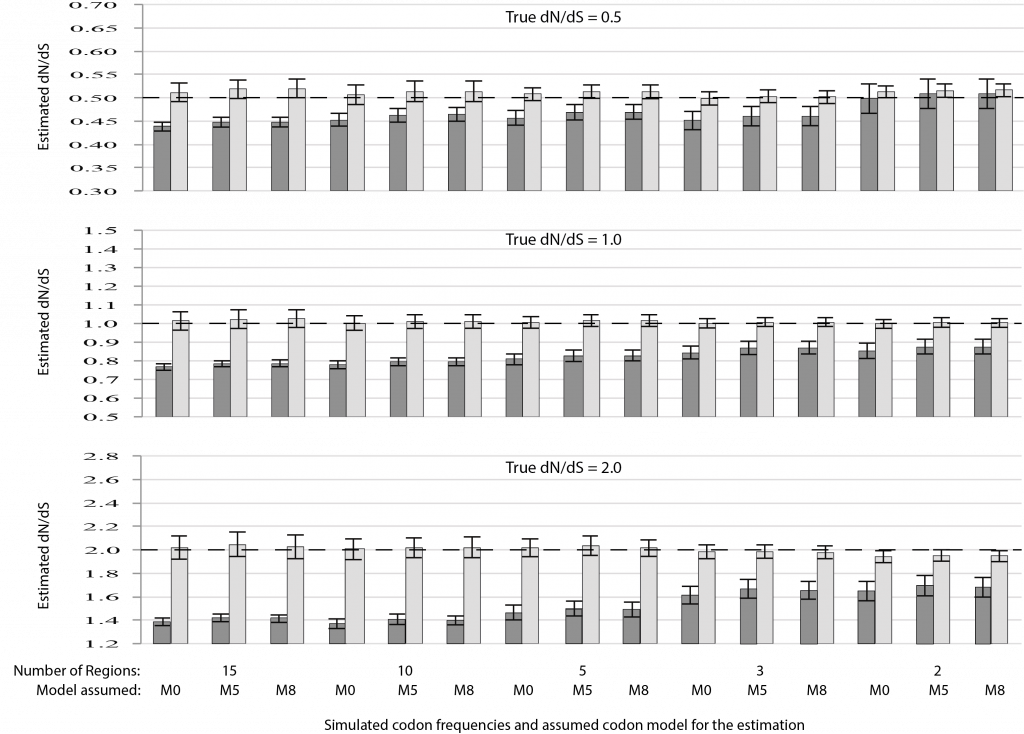
ProtASR2: Ancestral reconstruction of protein sequences accounting for folding stability
We developed a new version of our program «ProtASR» to infer ancestral sequences of proteins accounting for the protein folding stability. A methodology that produces more realistic ancestral sequences respect to those obtained using traditional empirical substitution models. This study was done in collaboration with Ugo Bastolla (Center for Molecular Biology Severo Ochoa). Just published in Methods in Ecology and Evolution, https://besjournals.onlinelibrary.wiley.com/doi/abs/10.1111/2041-210X.13341
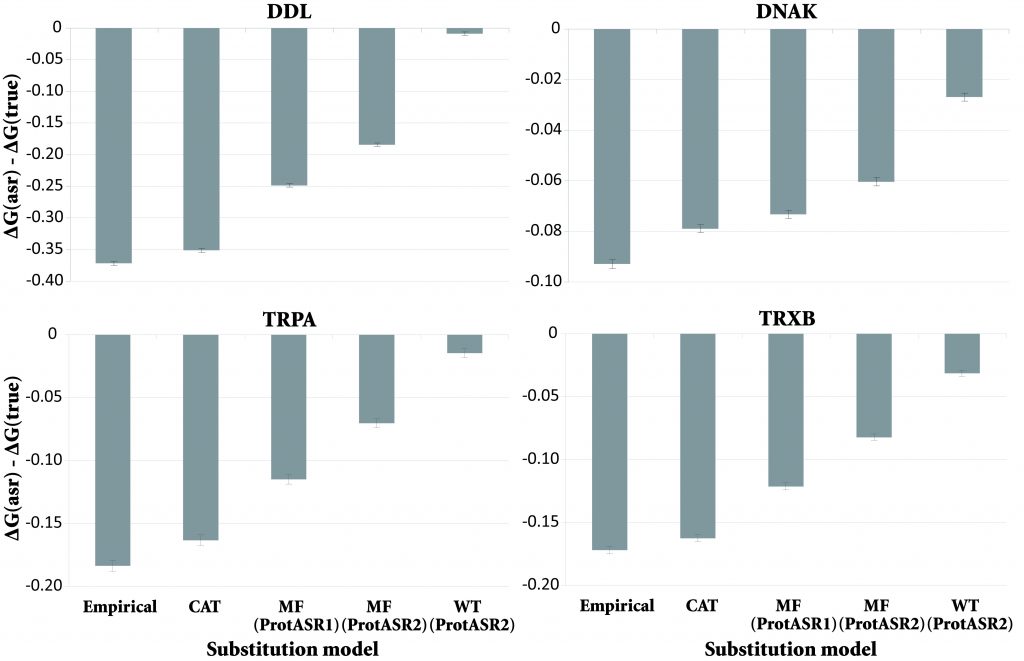
The Early Peopling of the Philippines based on mtDNA
We investigated the peopling of the Philippines by modern humans, including the estimation of evolutionary parameters and migration patterns such as migration corridors connecting the continent with the archipelago and the presence of long-distance dispersal. This study was done in collaboration with the lab of Antonio González-Martín (University Complutense of Madrid). Just published in Scientific Reports,
https://www.nature.com/articles/s41598-020-61793-7